BRAHMS Simulated Data Reconstruction
for Dummies!
This is not offiical method since official method does not
exist. However, this page will help you to run full GEANT
and reconstruct them as if it is a real data.
You are going to make cdat, ltr, gtr, dst files in order.
All of programs can be found in ~hito/analysis directory.
I assume that you have working brag and brat in your path.
I also assume that you have model input files for GEANT for brag.
How to Run BRAG (BRAHMS Geant Simulation) for full reconstruction
(FS+MRS+Global)
- You will find SubmitGeantSpect.rb in ~hito/analysis directory
- In this file, pay particular attention to
SingleGeantJob::createExec method. In this method, you will need
to setup all detectors according to the run of your choice. In
particular, look carefully at "set_tree" and "set_param" command and
change if it is needed. The figure below shows the setting used
for RUN04
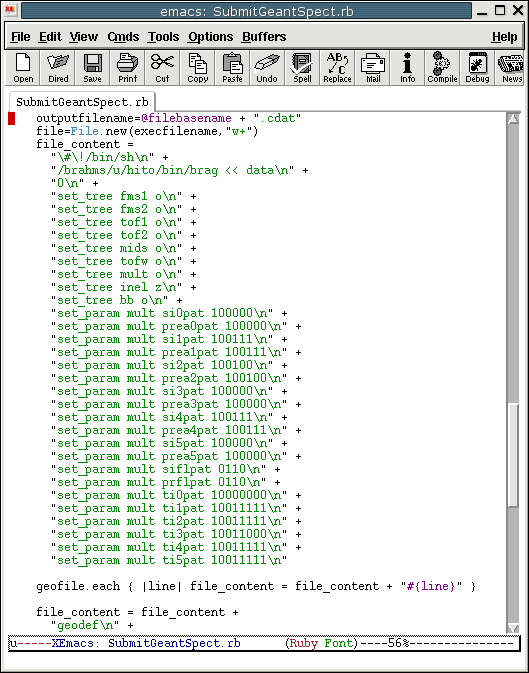
- Make a local directory for output of BRAG program. Every
rcas machine has /dataX directory. Just create your own
directory, eg. /data0/hito (NOTE: Don't place brag output
in your user area or our data disk area since brag output will quickly
will any aviable spaces.)
- Go to that directory, eg . cd /data0/hito
- Copy gbr2c.f from brag/src/user directory to this directory
- Run SubmitGeantSpect.rb by "ruby
~hito/analysis/SubmitGeantSpect.rb --runNumber=RunNo --cluster=crs(or
cas) --nevt=#Events --nEvtInFile=#EventsInAInputFile(see the
explanation) --startSeqNo=# --inputOpt=InputFile(use regular
expression"
- The reason for "nEvtInFile" as well as "nevt" is to avoid 2GB
file size limit problem. BRAG will fail if the output file(cdat)
exceeds 2GB because of OS (or g77) limitation. For example,
from one input file of 1GB with 10000 events, your total BRAG output
could be 10GB. Since brag output can not exceed 2GB, this
will fail. To go around this, you will need to use "skip" command
of brag. In this example, if you run 1000 events from 10000
events (1/10th), you have 1GB output. Then, you would set --nevt=1000
--nEvtInFile=10000 This will make 10 separate GEANT jobs,
each of which will analyze 1000 events starting from different section
of event file.
- About inputOpt option, to accomadate any kinds of names,
you can use Regular expression. For example, your geant input
directory is located at "/brahms/data21/scratch/ebj/BFlowSData/". And,
the format of your files are
"bflowsdata_phobv1v2linpt_v2_sNUMBER.zdat". You want to analyze
only "NUMBER" in the range of 10-19. Then, you would do
"/brahms/data21/scratch/ebj/BFlowSData/bflowsdata_phobv1v2linpt_v2_s1[0-9].zdat"
- NumNo is the acutal run number that the settings you are
interested in.
- As a comple example: If, in the above example, each file has
10000 Events and I wan to divide each file by 4 to reduce the output
file size, it will be "ruby ~hito/analysis/SubmitGeantSpect.rb
--runNumber=10438 --cluster=cas --nevt=2500 --nEvtInFile=10000
--inputOpt="/brahms/data21/scratch/ebj/BFlowSData/bflowsdata_phobv1v2linpt_v2_s1[0-9].zdat"
--startSeqNo=0" This will create 40 output files with format of
"simrunRunNoseqABCDE.cdat" at your starting directory. ABCDE will
be automatically incremented for each required output file.
- Now, you are done. This will take very long time. Go
get coffee, go joggin, follow by beer and sleep. By the time you
wake up in the next day, it might be done. Check your status by
"condor_q" or "condor_status"
<>
How to make simulated ltr file
- Go to the directory (and machine) containing your BRAG output
file (cdat file).
- You will use ~hito/analysis/SubmitGeantJob.rb file.
- Pay particular attention to bratmainScipt statement. You might
want to use your own scipt. The script will work for RUN04 AuAu,
but it may not work for other runs. It uses
~hito/analysis/Digitize.C in my case. (see the figure)
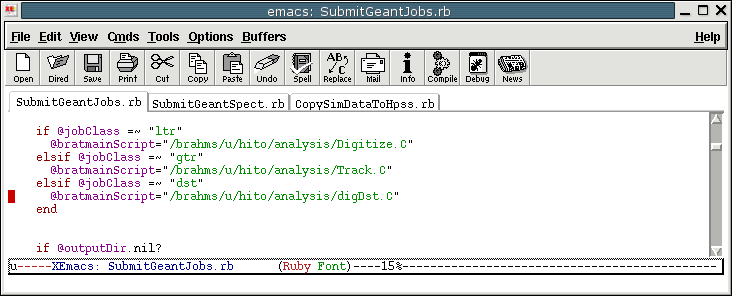
- Find/make the location for ltr files. Keep the same
directory structure as the offical data reconstruction. For
example, for run 04,
/brahms/data13/scratch/hito/data/run04/auau/200/rRunNumber/ltr
In this example, anything after hito is the same as official naming
format.
- Run the script by ruby ~hito/analysis/SubmitGeantJobs.rb
--cluster=crs(or cas) --runNumber=RunNo --jobClass=ltr
--outputDir="/brahms/data13/scratch/hito/data/run04/auau/200/rRunNo/ltr/"
--inputDir="/data0/hito/" --startSeq=startSequenceNo
--endSeq=EndSequenceNo
- startSeq and endSeq must correspond to input cdat data files of
your choice (NOTE: Don't submit more than 30 or so at a
time. Since it will copy each local cdat file to the temporary
location in the remote condor machine, it will severly limit the
disk/network bandwidth of the machine. By submitting more jobs,
it will slow down your process. This script will wait 30 seconds
between each sequence of input files.)
- Now, you are done. This will be done very
quickly. (Just keep an eye for every 15 minutes or so.)
How to make simulated gtr file
- You will use the same ~hito/analysis/SubmitGeantJob.rb file.
And, you can run from any locations as long as input and output are
/brahms/dataXY disks.
- Pay particular attention to bratmainScipt statement. You might
want to
use your own scipt. The script will work for RUN04 AuAu, but it
may
not work for other runs. It uses ~hito/analysis/Track.C in my
case.
- Find the location for gtr files. Keep the same directory
structure as the offical data reconstruction. For example, for
run 04,
/brahms/data05/scratch/hito/data/run04/auau/200/rRunNumber/gtr/
In this example, anything after hito is the same as official naming
format.
- Run the script by ruby ~hito/analysis/SubmitGeantJobs.rb
--cluster=crs(or cas) --runNumber=RunNo --jobClass=gtr --outputDir="/brahms/data05/scratch/hito/data/run04/auau/200/rRunNumber/gtr/"
--inputDir="/brahms/data13/scratch/hito/data/run04/auau/200/rRunNo/ltr/"
--startSeq=startSequenceNo --endSeqNo=EndSequenceNo
- Done. This will be done very quickly.
How to make simulated dst file
- You will use the same ~hito/analysis/SubmitGeantJob.rb
file. And, you can run from any locations as long as input and
output are /brahms/dataXY disks.
- ay particular attention to bratmainScipt statement. You might
want to
use your own scipt. The script will work for RUN04 AuAu, but it
may
not work for other runs. It uses ~hito/analysis/digDst.C in my
case.
- File the location for dst file. (This could be anywhere,
but, I suggest somewhere in /brahms/data21)
- Run the script by ruby ~hito/analysis/SubmitGeantJobs.rb
--cluster=crs(or cas) --runNumber=RunNo --jobClass=dst
--outputDir="/brahms/data21/scratch/hito/test/"
--inputDir="/brahms/data05/scratch/hito/data/run04/auau/200/rRunNumber/gtr/"
--startSeq=StartSequenceNo --endSeq=EndSequenceNo
- Done. This will be done shortly. It will make a singe
dst file. (unlike cdat, ltr,gtr files)
File Management
Since BRAG/BRAT makes require alot of data file space, it is necessary
to manage our disk space. Otherwise, you will find out that you
quickly run out of space. Therefore, you need to copy (or delete)
un-necessary files to HPSS archive. For this, you use
~hito/analysis/CopySimDataToHpss.rb It is meant to copy
cdat/ltr/gtr files. (since dst files are not large, it is not
included. You can copy them by hsi/htar command easily.)
- Log on to rcas machine as bramreco if you want to keep them as
official (recommended). Otherwise, you can logon as yourself.
- Go to the directory where descired files are located.
- ruby ~hito/analysis/CopySimDataToHpss.rb -r runNo -b beginSeqNo
-e endSeqNo -d dataType(run/ltr/gtr run for cdat files)
--inputDir="file directory"
- This program will copy files to ~bramreco (or
username)/sim/data/etc.... directory
- This program will also copy associated files: For cdat
file, it will copy all BRAG kumac file. For ltr/gtr, it will copy
histogram files.
- If you want to check the copied file, use "hsi/htar" command
- If you want to copy files from HPSS to desired directory, use
"hsi" command. For command for hsi, see Hsi home page. For htar
instruction, see htar
man page
- After confirming copy, you can delete unwanted files by shell
command (or any other ways you like)

